Frequently Asked Questions
On this page
The table of contents requires JavaScript to load.
Accounts and preferences
How do I get a CHPC account?
You can request to have a CHPC account here. To have a CHPC account, you need an active University of Utah uNIDand need to either be a PI or work with a PI.
If you work with a PI that does not have a CHPC account, your PI will need to apply for a CHPC account with the link above and mark in the application that they are a new PI.
If you do NOT have an active University of Utah uNID, please contact the CHPC at helpdesk@chpc.utah.edu so that we can request one for you.
How do I add a user to my CHPC group?
All users need a CHPC account to be added to your group. If the user does not yet have a CHPC account, please direct them to fill out a CHPC account application and select your group as their PI. Once the account is approved, it will automatically add the user to your group.
If the user does have an existing CHPC account, but is not a part of your group, you will need to email helpdesk@chpc.utah.edu, informing us that you would like to add that user to your group. We will require the user's name and uNID for this.
| Only PIs and delegates can request that a user be added or removed from a CHPC group. |
Being added to your CHPC group will allow the user to:
- Access your group's Slurm account and any allocated core hours you may have
- Access any group storage resouces that your group may have
I would like to change my shell (to bash or tcsh). How can I do this?
You can change your shell in the Edit Profile page by selecting the shell you'd like and clicking "Change." This change should take effect within fifteen minutes and you will need to re-login on any resources you were using at the time. That includes terminating all FastX sessions you may have running.
If you only need to use a different shell temporarily and don't want to fully change your own shell, you can initiate a new shell by invoking
it (e.g. with the tcsh or bash command, followed by pressing 'Enter') or you can pass commands as arguments to the
shell language (e.g. tcsh -c "echo hello").
I would like to change my email address. How can I do this?
You can change the email address CHPC uses to contact you in the Edit Profile page.
I would like to unsubscribe from CHPC email messages. How can I do this?
If you no longer work on any CHPC resources, you can unsubscribe by going to our unsubscribe page.
However, if you currently work on CHPC resources, please note that e-mail announcements are essential in updating the CHPC user base to changes in our systems and services and we strongly recommend to stay subscribed. We try to keep e-mails to a minimum, at most a couple a week, unless there is a computer downtime or other critical issue.
For ease of e-mail navigation and deletion, we have adopted four standard headlines for all of our messages:
- CHPC DOWNTIME - for announcements related to planned or unplanned computer downtimes
- CHPC PRESENTATION - for CHPC lectures and presentations
- CHPC ALLOCATIONS - for announcements regarding the CHPC resources allocation
- CHPC INFORMATION - for announcements different than the three above
Access and Connections
How do I connect to the CHPC clusters?
There are several options for connecting to CHPC systems:
- SSH directly to the cluster (addresses listed here, by cluster).
- Open OnDemand
- FastX
- 2 Windows systems, accessible via remote desktop: Beehive (in the general environment) and Narwhal (in the protected environment, for work with protected data).
I cannot login to a CHPC cluster.
There are multiple possibilities for a failure to login to CHPC systems:
It could be possible that the server for the cluster or node you are attempting to connect to is down, either due to planned or unplanned downtime or a localized failure of the login server.
If there is planned downtime, you would have received an email from the CHPC. If you did not receive an email, then make sure you have an up to date e-mail contact in your CHPC Profile in order to get our announcements, or, check our latest news.
In the case of a localized failure of the login node (e.g. the node you are immediately connected to when accessing CHPC systems), please attempt to connect to another CHPC node (e.g. kingspeak2 instead of kingspeak1, or a different cluster altogether). If the error persists, please make us aware by contacting us at helpdesk@chpc.utah.edu.
Your home directory is over quota (if trying to access CHPC systems from OnDemand OR FastX)
If you are attempting to connect to CHPC systems through the FastX software or through Open OnDemand, and your login attempts are failing, the most likely culprit is your home directory being over quota. This is due to the fact that both FastX and Open OnDemand write to your home directory.
To fix this, you need to ssh to CHPC clusters (without FastX) and clean up your home directory.
Another common possibility is a lockout after multiple failed login attempts due to incorrect passwords.
With this, your lockout could be initiated by CHPC systems OR the UofU campus.
In the case of lockout from CHPC systems:
Both CHPC and the UofU campus implement various measures to prevent "brute force" login attacks. After a certain number of failed logins within a specified time period, logins will be disabled for a small period of time. These time parameters are not made public for security reasons and vary between the general environment, protected environment, and the campus authentication, but, in general, the period of disablement is an hour or less from the last failed login attempt.
The best approach to deal with this is to try to log into a different machine (e.g. kingspeak2 instead of kingspeak1, or a different cluster altogether) or wait until login is enabled again on the login node/cluster you were first attempting to access. It is counter-productive to try to login again to the same login node, even with the correct password, as the disablement timer will reset.
In the case of lockout from UofU campus:
To check if campus authentication works, try any of the CIS logins. If you cannot successfully login to CIS, contact the campus help desk and inform them on the AD SSH lockout and ask them to unlock you.
If none of the above solutions work:
Contact the CHPC and make them aware of your login issues at helpdesk@chpc.utah.edu.
I can't ssh to a CHPC machine anymore and received a serious-looking error that includes "POSSIBLE DNS SPOOFING DETECTED" or "REMOTE HOST IDENTIFICATION HAS CHANGED"
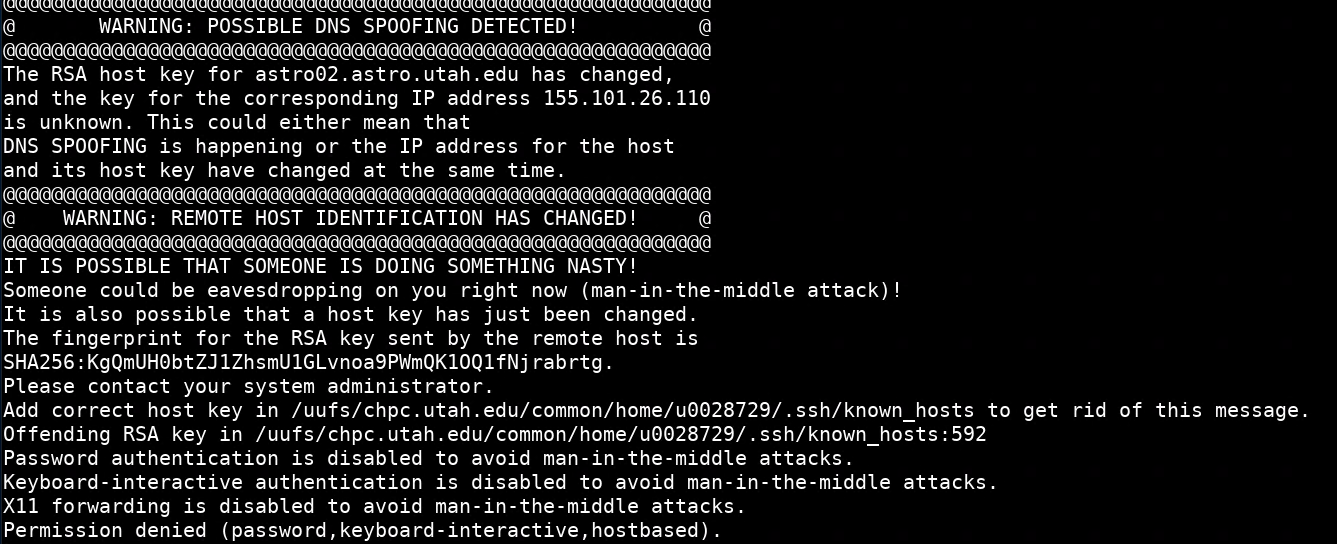
While this message looks scary, this error is usually benign. It occurs when the SSH keys on the machine you are trying to connect to change, most commonly with an OS upgrade on CHPC systems. There are two methods to get rid of this message and log in:
- open the file ~/.ssh/known_hosts in a text editor and delete the lines that contain the host name you are connecting to OR
- use the
ssh-keygencommand with a -R flag to remove the ssh keys for the given host
e.g. ssh-keygen -R kingspeak1.chpc.utah.edu
On the subsequent ssh connection to the machine should say something like the message below and let you login:
Warning: Permanently added 'astro02.astro.utah.edu,155.101.26.110 (ECDSA) to the list of known hosts
I am getting the message "Disk quota exceeded" when logging in.
By default, CHPC home directories provide 50GB of free storage. When the 50GB limit is exceeded, we prevent users from writing any more files to their home directory.
To display quota information, you can either run themychpc storage command inside a terminal session or log on to portal.chpc.utah.edu. For themychpc storage output, focus on the /home/hpc or the /home/chpc line.
As some access tools like FastX and Open OnDemand rely on storing small files in user's home directory upon logging in, access to these tools will fail.
If your storage is over 50GB, delete or move unnecessary files. To do that, log in using using ssh in a terminal tool,
such as Windows terminal, putty or Git bash, or terminal on a Mac or Linux machine, e.g ssh uNID@notchpeak1.chpc.utah.edu
To find what files/directories are taking the most space in your home directory, run
the ncdu command in your home directory to show disk space used per directory (the largest
directory will be at the top).
Please note that mychpc storage may not reflect the change in storage usage immediately. However, if one removes
enough data to get under quota, one will be able to write again, even if the report
still shows being over the quota.
To keep your file permanently, explore other storage solutions at CHPC.
If you clean a few files and are able to open FastX session again, a graphical tool
baobabis similar to ncdu but shows the usage in an easier-to-comprehend graphic.
I am running a campus VPN on my home computer and can't connect to certain websites.
Virtual Private Network (VPN) makes your computer to look like it currently resides on the University of Utah campus, even if you are off-site. However, depending on how the campus VPN is set up, you may not be able to access certain off-campus internet resources.
We recommend to only use VPN if one needs to:
- map network drives to CHPC file servers
- use remote desktop to connect to CHPC machines that allow remote desktop (e.g. Windows servers)
- connect to the Protected Environment resources
- ssh or FastX to connect to CHPC general environment Linux clusters
- access to the general environment Open OnDemand
All other resources do not need VPN. These include:
- accessing secure websites that require University authenticated login, such as CHPC's webpage, various other campus webpages (Canvas, HR,...) Box, Google Drive, etc.
High-Performance Computing Resources
How do I get an allocation?
General Environment Allocations:
An allocation is needed at the CHPC to work on our Granite and Notchpeak clusters. The CHPC awards allocations on our systems based on proposals submitted to a review committee. Research groups must submit requests for allocations. Please refer to our Allocation Documentation and our Allocations Policy for detailed information.
Please note that allocation requests must be completed by the CHPC PI or a designated delegate. To specify a delegate, the CHPC PI must email helpdesk@chpc.utah.edu and provide the name and uNID of the user you wish to have allocation delegate rights.
Protected Environment Allocations:
The protected environment has a separate allocation process; see the protected environment allocation information page for details. An allocation is required to work in Redwood, the Linux PE cluster.
I am getting a violation of usage policy warning from Arbiter that is confusing.
We monitor usage of the CHPC cluster login nodes as they are a shared and limited resource.
Our monitoring system, Arbiter, sends warnings if a user's CPU or memory usage exceeds limits on our login nodes. Even if one does not run time consuming calculations, sometimes applications such as FastX or VSCode Remote SSH puts enough load on the system to trigger these warnings.
In the email you received from Arbiter, there will be a section detailing the high-impact processes that caused Arbiter to trigger, such as the one below.
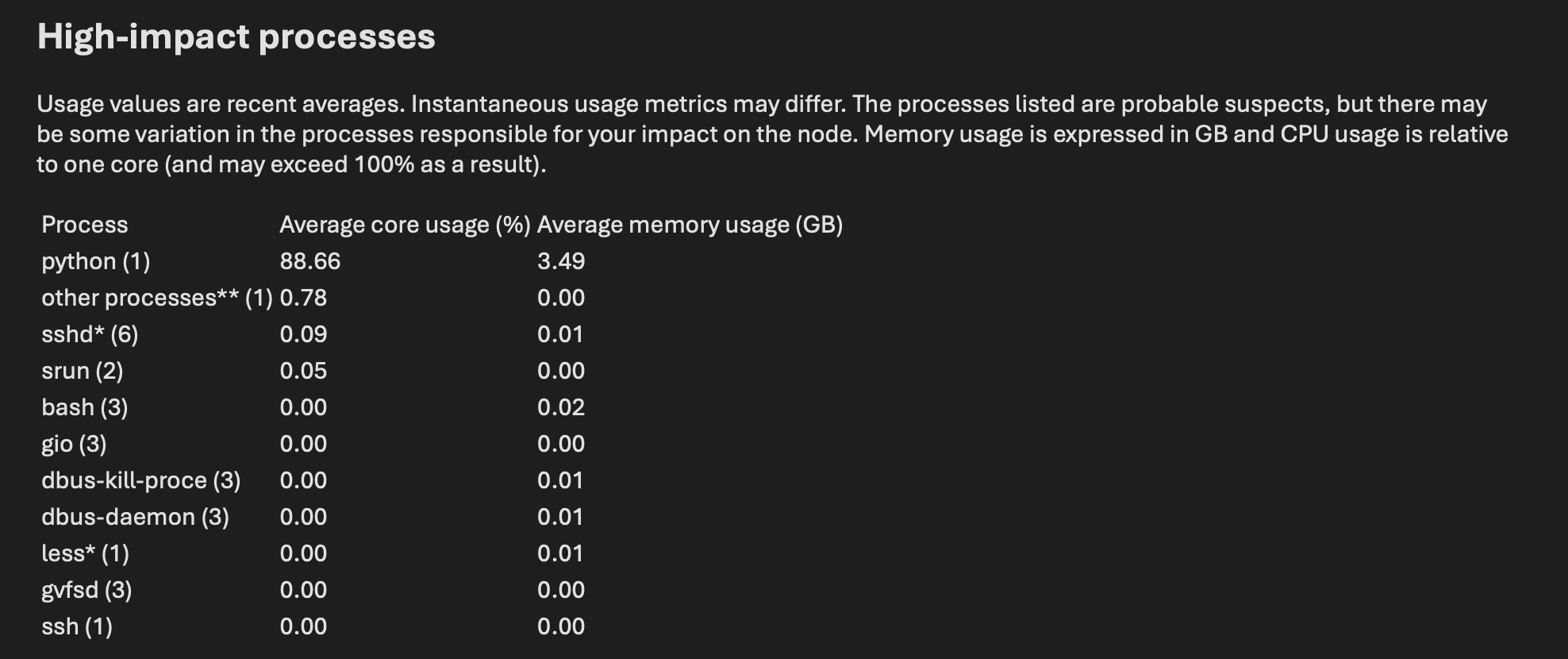
In the example above, you can see that the user was running a Python process that exceeded the limits we place on our login nodes. At this point, it would be appropriate for the user to terminate any Python process running on the login node and start the same job in an interactive session or on a compute node via Slurm.
What does this notice mean?
A high-performance computing (HPC) cluster consists of many computers, known as nodes. Most of the nodes—representing the vast majority of computational resources in a cluster—are compute nodes, which can be used for large or long-running computations. Other nodes include the login nodes: login nodes are a shared resource that all CHPC users can use to edit files, develop and debug software, and request computational resources. They are, in effect, the front doors to an HPC cluster; they are not where you should run large calculations. You are receiving this message because you are running a large or long-running task on a login node, also known as an interactive node. Running large or long-running calculations on login nodes can affect the responsiveness of the nodes for other CHPC users. It is required that you use compute nodes for your computations at the CHPC.
To use compute nodes, you will need to interact with the scheduler, Slurm. Slurm allocates resources by accounting for availability and the size of your job; it considers the number of CPU cores and quantity of memory required, for instance. We have documentation about Slurm on the CHPC website, and we recommend attending our introductory Slurm presentations if you haven’t used an HPC cluster before or would like a review.
How can I stop my processes on the login node?
Stopping processes that are consuming significant resources on the node should address the issue and prevent further emails.
- Log in to the node
Please note that you will not be able to see or stop the relevant processes from another node; the processes are specific to a node. If you are getting emails about usage on granite1.chpc.utah.edu, for example, you will need to log in to granite1.chpc.utah.edu, not granite2.chpc.utah.edu or granite.chpc.utah.edu (which could connect you to either granite1 or granite2). If you suspect it is your FastX session that triggered Arbiter, please log in to the affected node using FastX and terminate the FastX session. Keep in mind that FastX sessions stay alive after closing the session and need to be terminated explicitly. - Identify the processes that are using significant resources
You can use the commandps -u $USER -o pid,etime,%cpu,%mem,commto identify the process ID (PID) of each process. - Use the
killcommand with a process ID, likekill 1234567, to stop a process
If this does not work, trykill -9with the process ID, likekill -9 1234567; this is useful if processes are unresponsive or will not terminate.
I don't think I'm running anything on the node. What could be causing this?
Some processes may continue running for some time, even if you’ve logged out of the node and don’t believe you have any processes running. Please see the instructions for finding and stopping processes above.
If you are still unsure what is triggering the Arbiter warnings, please contact us at helpdesk@chpc.utah.edu.
I don't see my group space in a graphical file browser or when trying to use tab completion from the command line. Where did it go?
We do not keep group spaces mounted permanently. Instead, group spaces are automatically mounted with autofs when users access the group space.
Because of the use of autofs, group spaces may not immediately appear in file browsers or when using tab completion if the group spaces have not been accessed recently.
To auto-mount your group space, either list the contents of your group space or change
directories to your group space by using the full name from the command line (for
example, to get to my-group1 from your home directory, you can use cd ~/../my-group1). This will tell the system to mount your group space, and it should then be available
from file browsers and through tab completion.
If this does not work, please contact us at helpdesk@chpc.utah.edu.
I don't understand what the different Slurm accounts, partitions, and qualities of service (QoS) mean. What do I need to know to submit my job?
While the CHPC has different clusters that you can submit jobs to, the CHPC has set different Slurm partitions and qualities of service (QoS) to allow users access to a variety of different compute resources to meet their research needs. A Slurm Account and Quality of Service (QoS) will map to a particular partition, which is a group of compute nodes with similar features, i.e. whether or not they have CPUs or GPUs, if they are owned by a particular PI, or some other grouping mechanism.
By default, all partitions allow for node sharing, which allows for multiple jobs to run on one node. If you do not specify the number of cores (--ntasks) or amount of memory (--mem) required, you will receive a default amount. To request a whole node's resources, set --mem=0.
Please note that, below, <cluster> refers to any of the CHPC clusters (currently Granite, Notchpeak, Kingspeak, Lonepeak, and Redwood).
You can list the partitions (and their matching Slurm accounts and QoS') with the command 'mychpc batch'. If you are unsure which partition to choose, please use our handy tool for picking which partition is right for you.
- Partition: <cluster>
- Only contains CPUs.
- Partition: <cluster>-guest
- Only contains CPUs.
- This partition allows for jobs to be run on owner nodes.
- Jobs run on owner nodes are subject to preemption.
- Partition: <cluster>-gpu
- Contains CPUs and GPUs.
- To use a GPU, add --gres=gpu to your Slurm command.
- Partition: <cluster>-dtn
- This partition is connected to our Data Transfer Nodes and is useful for transferring large amounts of data at fast speeds, high performance, and low latency.
- Do not run computational jobs on the dtn nodes. They are for data transfer only.
- Partition: <cluster>-freecycle
- Your job should run on the freecycle partition only if you ran out of an allocation for the Granite, Notchpeak, or Redwood clusters.
- Freecycle jobs do not require an allocation, but are subject to preemption.
My Slurm job won't start.
There are many reasons why your Slurm job may not have begun. Here are a common list of reasons that may have prevented your Slurm job from starting:
-
Batch job submission failed: Invalid account or account/partition combination specified
This error message can indicate one of three things: 1) you supplied the incorrect account name or account/partition information to Slurm. 2) You are attempting to run a job on the notchpeak cluster, but your research group does not have an allocation or has used all of their allocation for the current quarter. 3) You are submitting the job on one cluster, but requested a partition on a separate cluster.
Solution to 1) If you suspect that you supplied the incorrect account name or account/partition information to Slurm, you can check your allocation with the
mychpc batchcommand. The output to this command will give you the correct account and partition combination for each Slurm partition that you have access to. Check the spelling in your batch script or interactive command and be sure you have access to the account and partition that you are requesting.
Solution to 2) To view your allocation status, and if you have run out of allocation for the current quarter, see this page. If your group is either not listed in the first table on this page or there is a 0 in the first column (allocation amount) your group does not have a current allocation. In this case, your group may want to consider completing a quick allocation request to supply your group with a small allocation for the remainder of the quarter. To receive a larger allocation for future quarters, you will need to submit an allocation request before the next deadline.
If you ran out of an allocation for the quarter, you must run your job either on our kingspeak or lonepeak clusters (if you need the general environment), on owner nodes, or on the freecycle partition. Jobs on the freecycle partition will have lower priority and will be preemptable.
Solution to 3) Either update the partition, account, and qos combination requested or use the -M
flag in your sbatch command. E.g. if you are logged into the Notchpeak cluster, but
are using the kingspeak partition, you can submit your Slurm script (called MySlurmScript.sh
in this example) with this command: sbatch -M kingspeak MySlurmScript.sh
-
Batch job submission failed: Node count specification invalid
The number of nodes that can be used for a single job is limited; attempting to submit a job that uses more resources than available will result in the above error. This limit of nodes one can request is approximately 1/2 the total number of general nodes on each cluster (currently 32 on notchpeak, 24 on kingspeak, and 106 on lonepeak). Detailed information on resources available for each cluster can be found here.
The limit on the number of nodes can be exceeded with a reservation or QOS specification. Requests are evaluated on a case-by-case basis; please contact us at helpdesk@chpc.utah.edu to learn more.
-
Required node not available (down, drained, or reserved) or job has "reason code" ReqNodeNotAvail
This occurs when a reservation is in place on one or more of the nodes requested by the job. The "Required node not available (down, drained, or reserved)" message can occur when submitting a job interactively (with srun, for instance); when submitting a script (often with sbatch), however, the job will enter the queue without complaint and Slurm will assign it the "reason code" (which provides some insight into why the job has not yet started) "ReqNodeNotAvail."
The presence of a reservation on a node likely means it is in maintenance. It is possible there is a downtime on the cluster in question; please check the news page and subscribe to the mailing list via the User Portal so you will be notified of impactful maintenance periods.
How do I know which cluster to submit my jobs to?
To answer this question, first answer: does your job belong in the general environment or protected environment (PE)? If working with Protected Health Information (PHI) connected with an IRB, it belongs in the PE on our redwood cluster.
If your job belongs in the general environment, you can submit to any of our three general environment clusters - notchpeak, kingspeak, or lonepeak - depending on your allocation. Notchpeak requires an allocation, while kingspeak and lonepeak do not require allocations.
You can view how busy each of our clusters are in the 'System Status' column to the right of the CHPC Homepage. If your job belongs in the general environment, we recommend submitting your job to the least busy cluster to reduce wait time.
My calculations or other file operations complain that the file can't be accessed, or it does not exist, even though I have just created or modified it.
This error may have many incarnations but it may look something like this:
ERROR on proc 0: Cannot open input script in.npt-218K-continue (../lammps.cpp:327)
It also occurs randomly, sometimes the program works, sometimes not.
This error is most likely due to the way the file system writes files. For performance reasons, it writes parts of the file into a memory buffer, which gets periodically written to the disk. If another machine tries to access the file before the machine that writes the file writes it to the disk, this error occurs. For NFS, which we use for all our home directories and group spaces, it is well described here. There are several ways to deal with this:
- Use the Linux sync command to forcefully flush the buffers to the disk. Do this both at the machine where the file writing and file reading occurs BEFORE the file is accessed. To ensure that all compute nodes in the job sync, do "srun -n $SLURM_NNODES --ntasks-per-node=1 sync ".
- Sometimes adding the Linux sleep command can help, to provide an extra time window for the syncing to occur.
- Inside of the code, use fflush for C/C++ or flush for Fortran. For other languages, such as Python and Matlab, google the language with "for flush" to see what options are there.
If neither of these help, please, try another file system to see if the error persists (e.g. /scratch/general/vast or /scratch/local), and let us know.
I can't access files from my group space or scratch in Jupyter or RStudio Server.
Unfortunately, both Jupyter and RStudio Server set one's home as a root of its file
system, so, one can't step "one directory down" and then browse back up to the group
file spaces.
There's a trick to get these portals to access the group space, or a scratch space
- create a symbolic link from that space to your home directory. To do this, in a
terminal window, run
ln -s /uufs/chpc.utah.edu/common/home/my-group1 ~/
Replace the my-group1 with the appropriate group space name. You'll then see the my-group1 directory in the root of your home and access it this way.
Similarly, for the scratch spaces, e.g. for /scratch/general/vast, we can do:
ln -s /scratch/general/vast/$USER ~/vast
My RStudio Server session does not start, or crashes on start.
Sometimes, the RStudio Server session files stored in user's home directory get corrupted, preventing starting new RStudio Server sessions. This is especially common if one chooses to automatically save the work space and doesn't terminate the RStudio Server session before deleting the job that runs it (e.g. in Open OnDemand).
Many of these situations can be prevented if one terminates the Open OnDemand RStudio Server session via its menu File - Quit Session, rather than just closing the web browser tab and deleting the job.
To remedy this situation, first try to remove the session files, by running
rm -rf ~/.local/share/rstudio/sessions/*.
If that does not help, move the whole RStudio settings directories.
mv ~/.local/share/rstudio ~/.local/share/rstudio-old.
Be aware that this will reset some customizations you may have done. If this does not work, move the user settings,
mv ~.config/rstudio ~/.config/rstudio-old .
Finally, if none of these steps work, contact our help desk. We have seen cases where the RStudio project files were saved in certain user home directories that were corrupted and they had to be removed by a member of the CHPC staff.
Starting the Emacs editor is very slow.
Emacs's initialization includes accessing many files, which can be slow in the network file system environment. The workaround is to run EMacs in the server mode (as a daemon), and start each terminal session using emacsclient command. The Emacs daemon stays in the background even if one disconnects from that particular system, so, it needs to be started only once per system start.
The easiest way is to create an alias for the emacs command as
alias emacs emacsclient -a \"\"
Note the escaped double quote characters (\"\"). This will start the emacs as a daemon if it's not started already, and proceeds to run in client mode.
Note that by default emacsclient starts in the terminal. To force start Emacs GUI, add the "-c" flag, e.g. (assuming the aforementioned alias is in place) "
emacs -c myfile.txt
Another solution, suggested by a user, is to add this line to your .emacs file in your home directory:
(setq locate-dominating-stop-dir-regexp "\\`\\(?:/uufs/chpc.utah.edu/common/home/[^\\/]+/\\|~/\\)\\'")
Opening files in Emacs is very slow.
We have yet to find the root of this problem but it's most likely caused by the number of files in a directory and the type of the file that Emacs is filtering through. The workaround is to read the file without any contents conversion:
M-x find-file-literally <Enter> filename <Enter>
After opening the file, one can tell Emacs to encode the file accordingly, e.g. to syntax highlight shell scripts:
M-x sh-mode <Enter>
To make this change permanent, open the ~/.emacs file to add:
(global-set-key "\C-c\C-f" 'find-file-literally)
My program crashed because /tmp filled up.
Linux defines temporary file systems at /tmp or /var/tmp, where temporary user and system files are stored. CHPC cluster nodes set up temporary
file systems as a RAM disk with limited capacity. All interactive and compute nodes
have also a spinning disk local storage at /scratch/local.
If a user program is known to need temporary storage, it is advantageous to set environment
variable, TMPDIR, which defines the location of the temporary storage, and point it to /scratch/local. Or, even better, create a user specific directory, /scratch/local/$USER, and set /scratch/local to that as shown in our sample at /uufs/chpc.utah.edu/sys/modulefiles/templates/custom.[csh,sh] .
How do I check whether my job is running efficiently?
You can check the efficiency of your jobs on portal.chpc.utah.edu.
You can also use command-line tools to view performance metrics. If your job is underperforming, one possibility is that you are not utilizing all of the available CPUs that you requested. To find this information, run the following command:
pestat -u $USER
Any information shown in red is a warning sign. In the example output below the user's
jobs are only utilizing one CPU out of 16 or 28 available:
Hostname Partition Node Num_CPU CPUload Memsize Freemem Joblist
State Use/Tot (MB) (MB) JobId User ...
kp016 kingspeak* alloc 16 16 1.00* 64000 55494 7430561 u0123456
kp378 schmidt-kp alloc 28 28 1.00* 256000 250656 7430496 u0123456
Another possibility is that the job is running low on memory, although we now limit
the maximum memory used by the job via Slurm so this is much less common than it used
to be. However, if you notice low free memory along with low CPU utilization, like
in the example below, try to submit on nodes with more memory by using the #SBATCH --mem=xxx option.
An example of pestat output of a high memory job with a low CPU utilization is below:
Hostname Partition Node Num_CPU CPUload Memsize Freemem Joblist
State Use/Tot (MB) (MB) JobId User ...
kp296 emcore-kp alloc 24 24 1.03* 128000 2166* 7430458 u0123456
My calculations are running more slowly than expected.
First, check how efficiently are your jobs using the compute nodes. This can give some clues as to what the problem is.
There can be multiple reasons for this, ranging from user mistakes to hardware and software issues. Here are the most common causes, in the order of commonality:
- Not parallelizing the calculation. In an HPC environment, we obtain speed by distributing the workload onto many processors. There are different ways to parallelize depending on the program workflow, starting from independent calculations to explicit parallelization of the program using OpenMP, MPI, or interpreted languages like Python, R or Matlab. For some basic information on how to use explicit parallelization, see our Introduction to Parallel Computing lecture or contact us.
- User does not supply the correct number of parallel tasks or hard coded the number of tasks to run instead of using Slurm variables like $SLURM_NTASKS or $SLURM_CPUS_PER_NODE. Check your Slurm script and program input files for this. If in doubt, contact us.
- Inefficient parallelization. MPI can especially be sensitive to how efficiently the program parallelization is implemented. If you need help in analyzing and fixing the parallel performance, contact us.
- Hardware or software issues on the cluster. If you rule out any of the issues listed above, please contact us.
I'm receiving a "Killed" or "OOM" error from Slurm.
The CHPC keeps strict memory limits in jobs through the Slurm scheduler. If your job ends prematurely, please check if the job output has "Killed" or "OOM" at or near the end of the output. That would signalize that the job was killed due to being low on memory.
Occasionally the Slurm memory check does not work and your jobs end up either slowing
down the nodes where the job runs or puts the nodes into a bad state. This requires
sysadmin interaction to recover the nodes, and we usually notify the user and ask
to correct their behavior - either by asking more memory for the job (with the #SBATCH --mem=xxx option), or by checking what they are doing and lowering their memory needs.
In either case, in these situations it is imperative to monitor the memory usage of
the job. A good initial check is to run the pestat command. If you notice that the memory is low, the next step would be to ssh to the affected
node and run the top command. Observe the memory load of the program and if it is high and you notice
kswapd processes taking some CPU time, this indicates that the program is using too much
memory. Delete the job before it ends up putting the node in a bad state, remedy the
memory needs as suggested above.
How can I use all processors in a multi-node job with different CPU core counts per node?
In some cases (e.g. when an owner has several generations of nodes) it is desireable to run a multi-node job that spans nodes with different CPU core counts. To utilize all the CPUs on these nodes, instead of defining the job's process count with the SLURM_NTASKS variable provided by Slurm, one has to explicitly specify the total CPU core count on these nodes.
This is achieved by specifying only--nodes, not--ntasks, in the node/tasks request part of the Slurm batch script. The second step is to
calculate how many CPU cores are available on all the job's nodes, which is stored
in the JOB_TASKS variable. This variable is supplied (e.g. to mpirun) to specify the
number of tasks.
#!/bin/bash
#SBATCH --nodes=12
....
JOB_TASKS=`echo $SLURM_JOB_CPUS_PER_NODE | sed -e 's/[()]//g' -e 's/x/*/g' -e 's/,/+/g'
| bc`
mpirun -np $JOB_TASKS program_to_run
My job needs access to GPU resources. How do I request a GPU node for my job?
via Slurm:
Add the following to your batch script:
#SBATCH --gres=gpu
You can alternatively request certain types and counts of GPU resources as such:
#SBATCH --gres=gpu:v100:4
The above request 4 v100 GPUs.
via Open OnDemand:
You can request access to GPUs when running software on Open OnDemand. When filling out the fields to request CHPC resources for your software, you will see a field called 'Advanced Options', such as below:
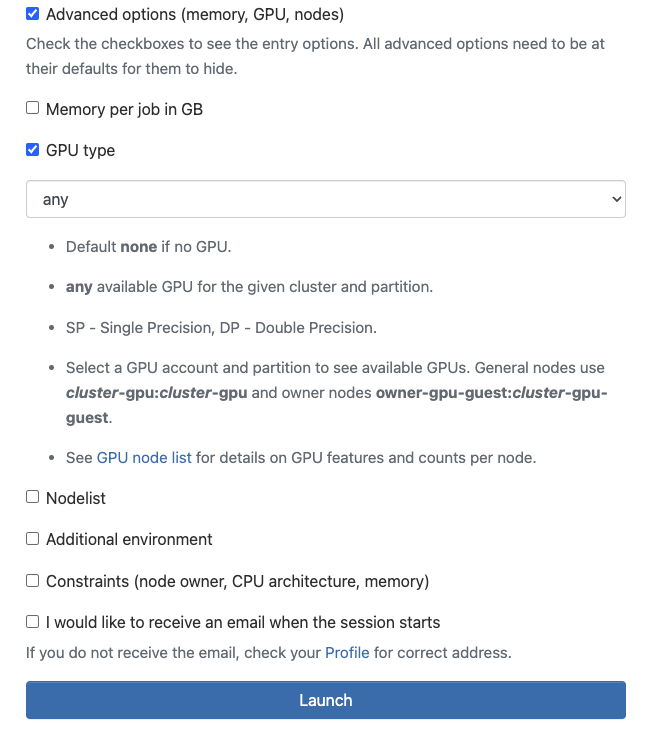
You must click on the 'Advanced Options' box for GPU options to appear. There, you can click 'GPU type' to request GPU resources and specify what types of GPUs you need for your job. Put 'any' if you just need any GPU.
**by not clicking advanced options and not specifying a GPU type, your Open OnDemand session will not have GPU resources.
My program crashes with error "error while loading shared libraries: … No such file or directory".
This error means that the program can't find shared libraries on which it depends. The full error message specifies the library name, e.g. "error while loading shared libraries: libnetcdf.so.19: cannot open shared object file: No such file or directory".
To get the program running, one needs to tell the program the path where the library
is, either by setting this path to the LD_LIBRARY_PATH environment variable, or by loading a module that has this library (which sets the
LD_LIBRARY_PATH). To list all the shared libraries that are needed by the executable,
use the ldd command, e.g. ldd myprogram.
If one builds a program from source, it's the best to modify the rpath of the program by adding linker flag -Wl,-rpath=PATH, PATH being the path where the library is located. For example, in the above mentioned
missing NetCDF library, one could achieve this by loading the gcc/8.5.0 netcdf/4.9.0 modules and in their link line or LDFLAGS, set -Wl,-rpath=$NETCDF_ROOT/lib. The NETCDF_ROOT environment variable is defined in the netcdf/4.9.0 module and points to the path where the libnetcdf.so is located.
I am getting an error like "libc.so.6: version GLIBC_2.33 not found" when trying to run a program.
This means that the program executable was built on a newer operating system, than what our clusters run. Different OS versions use different base OS library called glibc, which is backward, but not forward compatible. That is, programs built with newer glibc don't run on older OSes. In the error message above, glibc 2.33 is referenced. The Rocky Linux 8, which is at the time of this writing used at CHPC clusters, uses glibc 2.28, i.e. an older glibc than the program above requires.
If the program supplies binaries for different Linux OSes, try to get one that's close to Rocky Linux 8 / RHEL8 that we run. A list of Linux distributions and their respective glibc versions can be found here.
Another option is to see if the program can be easily built from the source natively on our systems, or with a package manager like Spack.
If other binary is not available, and building from the source is cumbersome, either look if this program is available in a container, or create a container that runs the OS on which this program was built on, and run it in this container. Contact us if you need help.
My RStudio or Jupyter session on OnDemand is not able to save files
This error occurs when the OnDemand session reaches the time limit of the authenticated session. This time limit is set by the campus in the general enviroment, observed at about 2 hours at the time of this writing. In the protected environment we limit this to 30 minutes.
If you get this error, reload the main OnDemand webpage, which will prompt login window followed by a new login. From this new login window, navigate to My Interactive Session and click the blue Launch button to open the existing interactive app session in a new browser tab. This new session will be in the same state where your old session was at the time when the authentication limit ran out. Close the old session's tab as this session has lost connection with the server and while it's responding, it's not authenticated anymore and as such can not save any files.
Is your question not on this list?
If our FAQ section did not answer your question(s), please email us at helpdesk@chpc.utah.edu and one of our dedicated team members will assist you.
